

for ex in tfds.load('cifar10', split='train'): They are all accessible in ourįor a quick introduction. In the current tensorflow-datasets package. Note: The datasets documented here are from HEAD and so not all are available
diabetic_retinopathy_detection (manual). "Error occurred when finalizing GeneratorDataset iterator: Failed precondition: Python interpreter state is not initialized. I believe the input matrix of the fit method should contain Image Index, height, widht, depth so it should have 4 dimensions while my x_train array only has 3 dimensions and doesn't have any dimension about the depth of the image. Model.fit(generator.flow(x_train, y_train, batch_size=32), steps_per_epoch=len(x_train)/32, epochs=epochs) 
I tried to use this code: generator.fit(x_train)
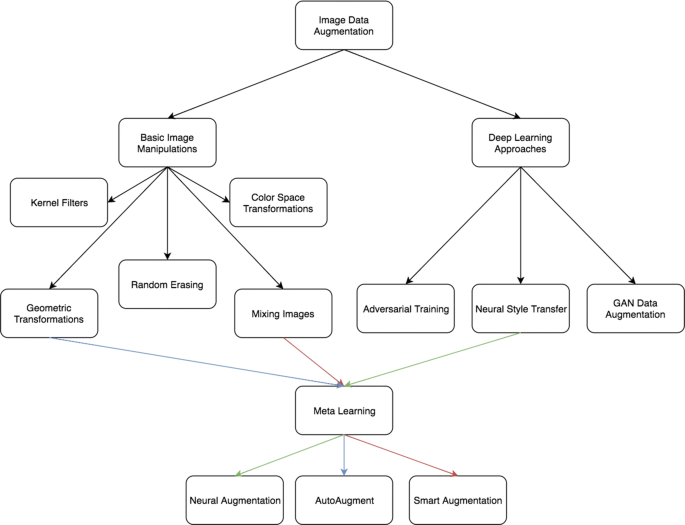
 What other options are there to prevent overfitting on mnist?. Does data augmentation even make sense with the mnist dataset?. If not: How can I train the model I've created with this generator? (or do I have to implement data augmentation in a completely different way?). Can I somehow convert the generator to data that I can use as parameters in the fit method of the model?. Here is the graph that I get from the training history: However, I don't know how to use this generator for the model I've created because the only method I've found was the fit method of the generator but I want to train my model and not the generator. Now I want to train the model with my train_model_with_data_augmentation function: train_model_with_data_augmentation( Then I create an ImagaDataGenerator: generator = tf.( I also use this function when I try to optimize the hyperparameters (hence the many parameters). The function returns a compiled but untrained model. Model.add(Dense(total_classes, activation='sigmoid')) Model.add(Dense(_dense_neurons, activation='relu')) This is the function definition: def create_model(_learning_rate=0.01, _momentum=0.9, _decay=0.001, _dense_neurons=128, _fully_connected_layers=3, _loss="sparse_categorical_crossentropy", _dropout=0.1): I create a model with a "create_model" function: untrained_model = create_model() Test_vector_labels = _categorical(test_labels, total_classes) Tr_vector_labels = _categorical(tr_labels, total_classes) #convert labels into the respective vectors #function which returns the amount of train images, test images and classesĪmount_train_images, amount_test_images, total_classes = get_data_information(tr_images, tr_labels, test_images, test_labels) Tr_images, test_images = preprocess(tr_images, test_images) (tr_images, tr_labels), (test_images, test_labels) = mnist.load_data() To combat this problem, I wanted to use data augmentation:įirst I load the data: #load mnist dataset The problem is that my model already overfits after 1 or 2 epochs. I want to train a keras neural network on the mnist dataset.
What other options are there to prevent overfitting on mnist?. Does data augmentation even make sense with the mnist dataset?. If not: How can I train the model I've created with this generator? (or do I have to implement data augmentation in a completely different way?). Can I somehow convert the generator to data that I can use as parameters in the fit method of the model?. Here is the graph that I get from the training history: However, I don't know how to use this generator for the model I've created because the only method I've found was the fit method of the generator but I want to train my model and not the generator. Now I want to train the model with my train_model_with_data_augmentation function: train_model_with_data_augmentation( Then I create an ImagaDataGenerator: generator = tf.( I also use this function when I try to optimize the hyperparameters (hence the many parameters). The function returns a compiled but untrained model. Model.add(Dense(total_classes, activation='sigmoid')) Model.add(Dense(_dense_neurons, activation='relu')) This is the function definition: def create_model(_learning_rate=0.01, _momentum=0.9, _decay=0.001, _dense_neurons=128, _fully_connected_layers=3, _loss="sparse_categorical_crossentropy", _dropout=0.1): I create a model with a "create_model" function: untrained_model = create_model() Test_vector_labels = _categorical(test_labels, total_classes) Tr_vector_labels = _categorical(tr_labels, total_classes) #convert labels into the respective vectors #function which returns the amount of train images, test images and classesĪmount_train_images, amount_test_images, total_classes = get_data_information(tr_images, tr_labels, test_images, test_labels) Tr_images, test_images = preprocess(tr_images, test_images) (tr_images, tr_labels), (test_images, test_labels) = mnist.load_data() To combat this problem, I wanted to use data augmentation:įirst I load the data: #load mnist dataset The problem is that my model already overfits after 1 or 2 epochs. I want to train a keras neural network on the mnist dataset.








 0 kommentar(er)
0 kommentar(er)
